Monocytes are a type of white blood cell that play crucial roles in the immune system. Monocytes exhibit significant variability in their appearance and functions, leading to heterogeneity in their roles within the body. By utilizing high-dimensional technologies, researchers gain a more comprehensive understanding of the different subsets of monocytes and their functions.
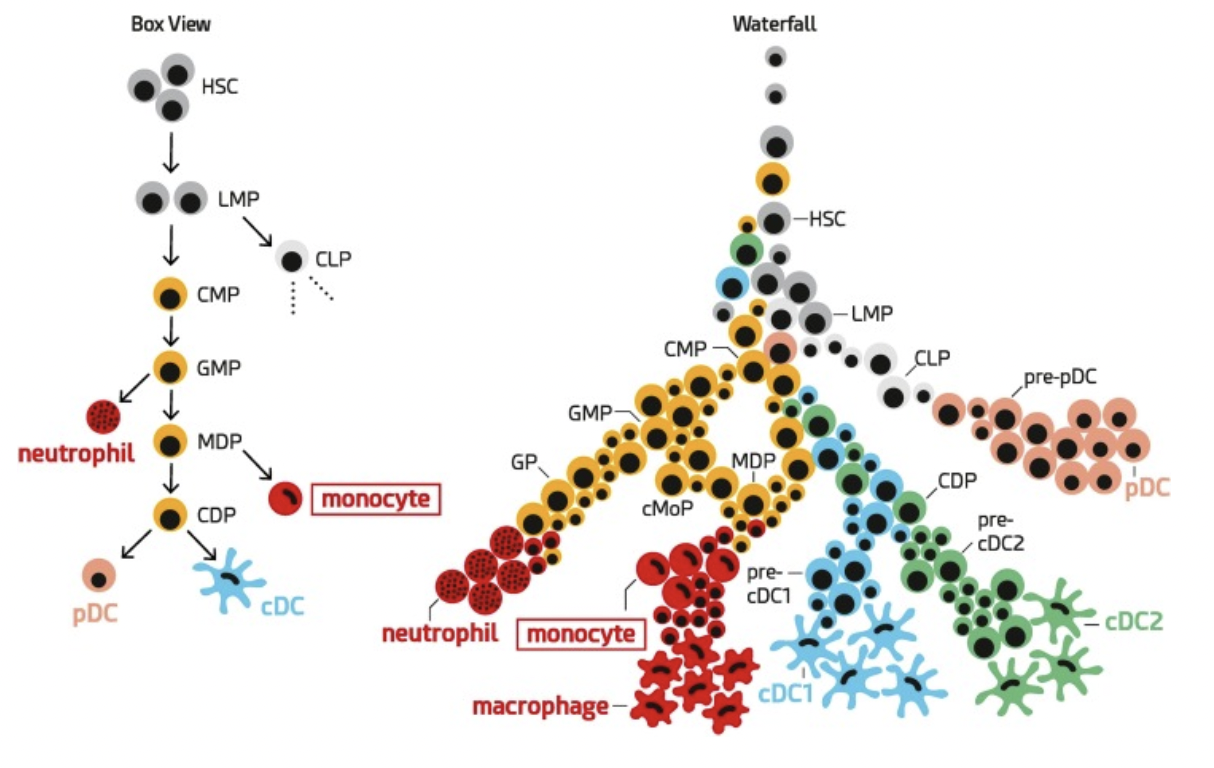
The rigid model of myeloid cell differentiation follows the idea that a particular type of precursor cell called CMP would give rise to another precursor called GMP, which could then become either a monocyte or a neutrophil or further differentiate into MDP and CDP, which are precursors of pDCs and cDCs (left).
Thanks to more recent advanced in single-cell comic techniques, we now understand that the cells follow a more fluid pathway. Multiple stages of progenitor cells gradually become more committed to specific cell types during the process (right).
In the context of cancer, observable characteristics and functions of monocytes lead to significant implications for disease progression. Understanding these changes can provide insights into potential therapeutic targets and strategies to manipulate monocyte functions to combat cancer.
Experimental workflow using CyTOF. Experimental setup for the processing of frozen PBMC from matched samples before and after PD-1 immunotherapy using metal-labeled antibodies and acquisition by mass cytometry.

Our article titled “High-Dimensional Analysis of the Immune Landscape during Anti-PD-1 Immunotherapy of Melanoma Predicts Responsiveness”, published in Nature Medicine in January 2018, explores the use of advanced analytical high-dimensional single-cell techniques to understand the immune response in melanoma patients undergoing anti-PD-1 immunotherapy and predict their responsiveness to the treatment.
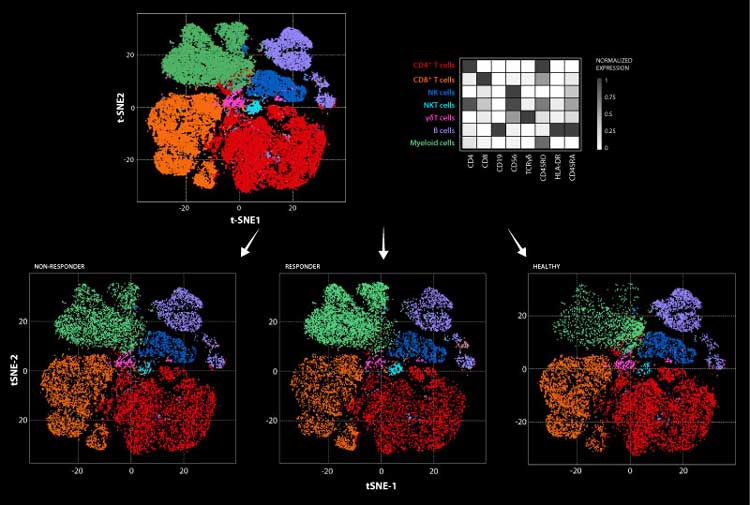
Immune instagrams were generated using the FlowSOM algorithm over millions of cells from patients from all groups. Cells are colored according to the cluster they were assigned by manual annotation using the exemplified heatmap. The heatmap represents the median arcsinh-transformed marker expression normalized to 0-1 range of respective markers within the 7 cellular clusters. In a top to bottom approach samples are then allocated to the different analysis groups: non-responders (NR), responders (R), and healthy donors (HD).
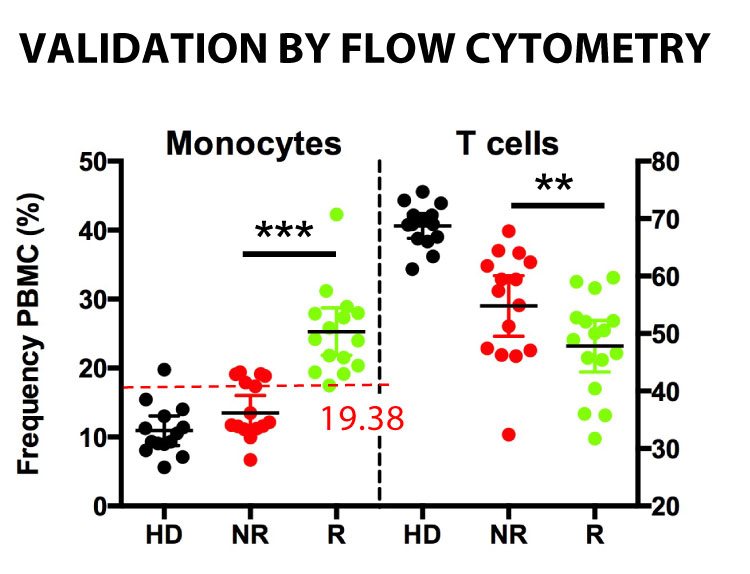
One of the study’s key findings is that specific monocyte subsets before treatment are associated with a higher likelihood of positive response to anti-PD-1 immunotherapy. This suggests that the composition of monocytes within the immune landscape can serve as a predictive marker for treatment outcomes.
Bar graphs showing individual patient sample composition before immunotherapy after extraction from the algorithm generated healthy donors (HD) non-responders (NR) and responders (R) clusters.

Identifying monocyte subsets as predictors of immunotherapy responsiveness has significant implications for melanoma treatment. It can help clinicians select patients who have a higher probability of responding positively to the therapy, optimizing treatment outcomes, and potentially avoiding ineffective or unnecessary treatments for patients who are unlikely to benefit.
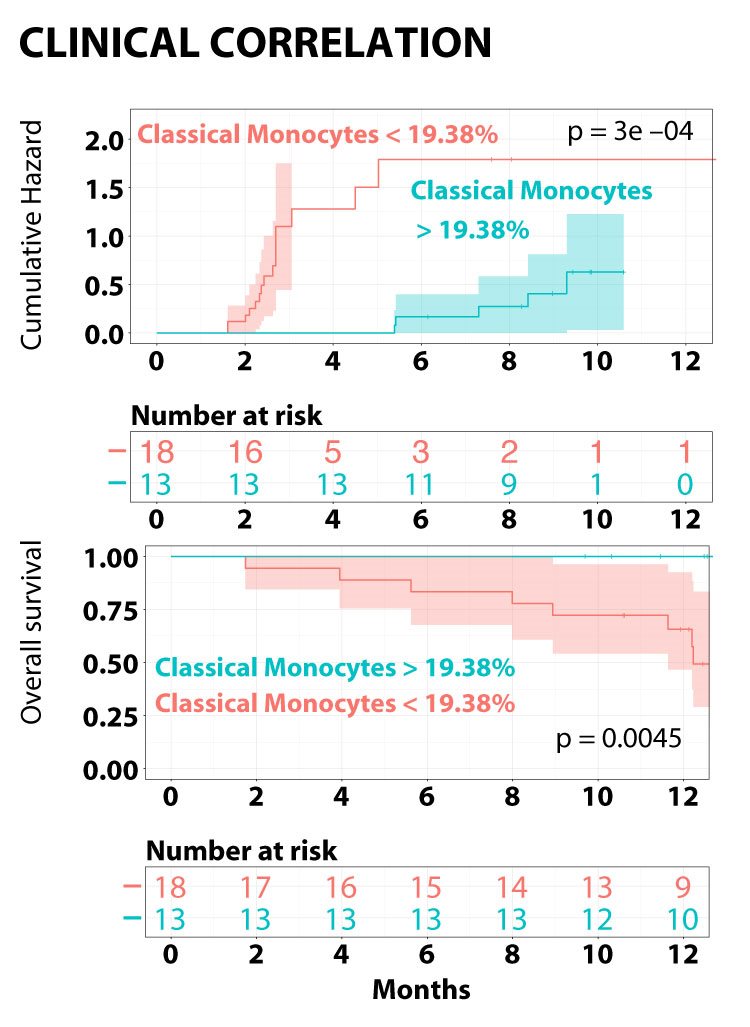
Classical monocyte frequencies above (blue) and below (red) 19.38% over time (months) correlate with cumulative hazard and overall survival in patients with.
We currently investigate which monocytes changes support anti-tumor immunity during immunothearpy.
Monitoring the immunome in patients treated with novel combination immunotherapy
Mass cytometry analysis of the immune cell response to a novel superagonistic IL-15 plus anti-PD-1 immunotherapy in patients with refractory non small cell lung cancer (NSCLC).
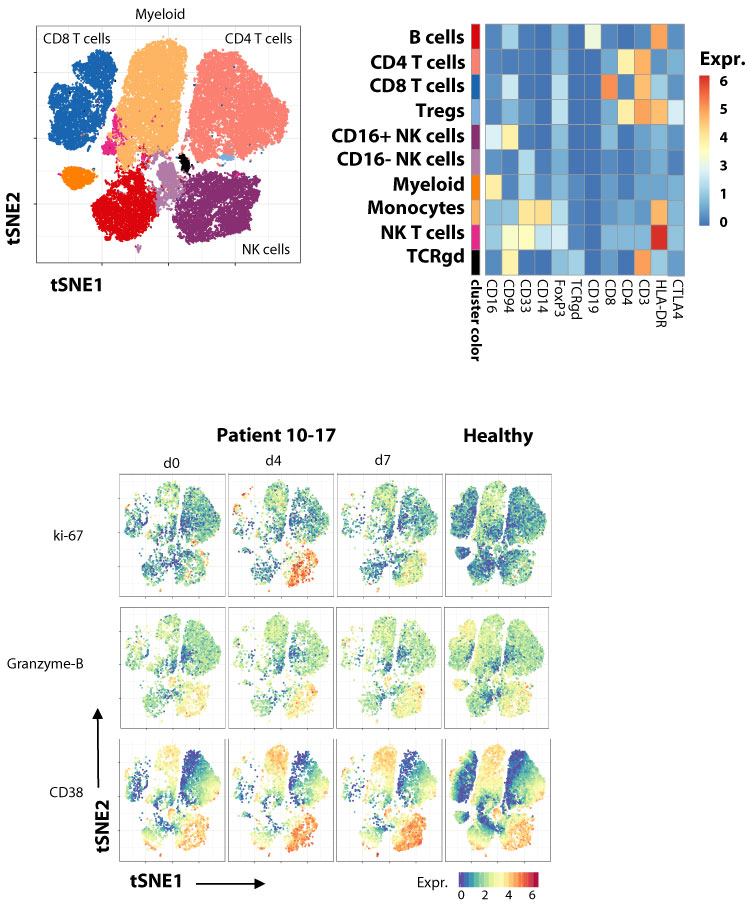
In the second article, published in April 2018 in Lancet Oncology, together with MUSC faculty members, first author John Wrangle and last author, Mark Rubinstein, we reported the first-in-human experience of an IL-2Rbetagamma agonist with a drug targeting the PD-1/PD-L1 pathway. This report demonstrates that these two classes of drugs can be administered safely, that there is evidence of clinical efficacy, and a comprehensive single cell high dimensional analysis of the immune profile in these patients.